library(tidyverse)
library(ggsci)
library(scales)
library(urbnmapr)
library(plotly)
theme_set(theme_minimal(base_size = 14))
Intro
Today, not a lot of text. Just the code and output for selected COVID graphs.
Load
covid <- read_csv("https://covid.ourworldindata.org/data/owid-covid-data.csv")
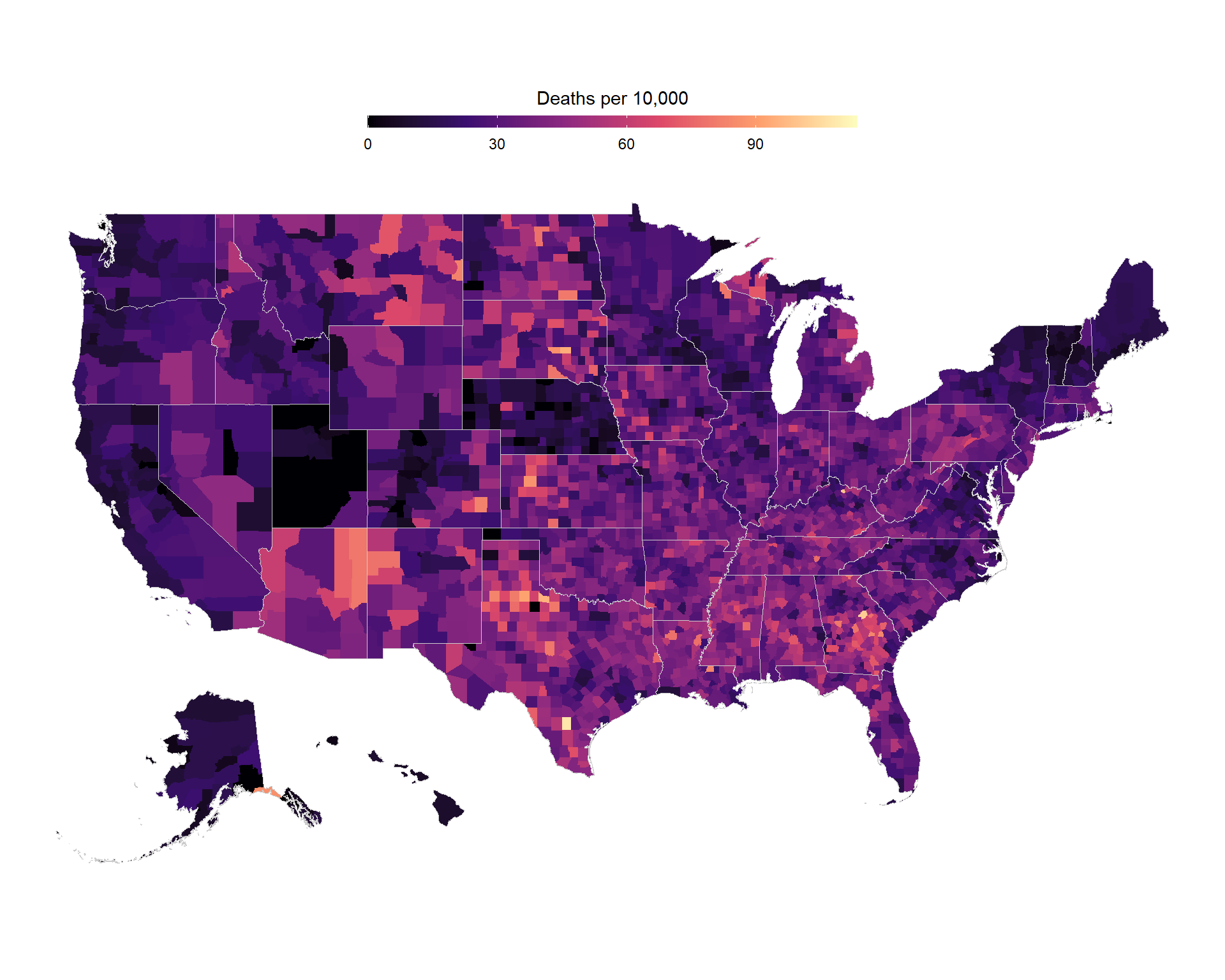
US Counties Map of Death Rates
us_deaths <- read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_US.csv")
counties %>%
mutate(county_fips = parse_number(county_fips)) %>%
left_join(
us_deaths %>%
select(FIPS, Population, deaths = last_col()) %>%
mutate(deaths_10000 = 10000 * deaths / Population),
by = c("county_fips" = "FIPS")
) %>%
ggplot(aes(long, lat, group = group)) +
geom_polygon(aes(fill = deaths_10000)) +
scale_fill_viridis_c(
option = "magma",
guide = guide_colorbar(
barwidth = 20,
barheight = 0.5,
title.position = "top",
title.hjust = 0.5,
title = "Deaths per 10,000"
)
) +
geom_polygon(data = states, color = "grey80", fill = NA, size = 0.2) +
theme_void() +
theme(legend.position = "top") +
coord_map() +
labs(x = "Longitude", y = "Latitude")
Time-series of Cases, Deaths, Vaccinations, and Tests
g1 <- covid %>%
filter(location %in% c("Israel")) %>%
select(
location,
date,
new_cases_smoothed_per_million,
new_deaths_smoothed_per_million,
new_people_vaccinated_smoothed_per_hundred,
new_tests_smoothed_per_thousand
) %>%
pivot_longer(-c(location, date)) %>%
mutate(name = str_to_title(str_replace_all(name, "_", " "))) %>%
replace_na(list(value = 0)) %>%
ggplot(aes(date, value, col = name)) +
geom_line() +
facet_wrap( ~ name, scales = "free_y", ncol = 1) +
scale_x_date(date_labels = "%b %Y") +
labs(x = "Date", y = NULL, title = "Israel, summary") +
scale_y_continuous(labels = comma_format()) +
theme(legend.position = "none")
ggplotly(g1)
Vaccination Rates by Continent
set.seed(123)
g2 <- covid %>%
select(continent, location, population, date, total_vaccinations_per_hundred) %>%
drop_na(total_vaccinations_per_hundred, continent) %>%
group_by(location) %>%
filter(date == max(date)) %>%
ungroup() %>%
mutate(continent = fct_reorder(continent, total_vaccinations_per_hundred),
Vaccinations = total_vaccinations_per_hundred,
Location = location) %>%
ggplot(aes(Vaccinations, continent, label = Location)) +
# geom_boxplot(aes(fill = continent), alpha = 0.3, outlier.alpha = 0, width = 0.5) +
geom_jitter(aes(fill = continent, size = population), width = 0, height = 0.25, shape = 21, color = "black") +
scale_fill_brewer(type = "div", palette = 5) +
scale_color_brewer(type = "div", palette = 5) +
theme(legend.position = "none") +
labs(y = NULL, x = "Vaccinations per hundred people") +
labs(title = "Inequality in Vaccination Rates") +
scale_size_continuous(range = c(3,8))
ggplotly(g2, tooltip = c("Location", "Vaccinations"))
Vaccinations Scatter-Plot
g3 <- covid %>%
select(continent, location, date, population,
people_vaccinated, people_fully_vaccinated) %>%
na.omit() %>%
group_by(location) %>%
filter(date == max(date)) %>%
ungroup() %>%
mutate(across(ends_with("vaccinated"), ~100 * .x / population)) %>%
filter(people_vaccinated <= 100,
people_fully_vaccinated <= 100,
people_fully_vaccinated <= people_vaccinated
) %>%
mutate(fully_vaccinated_rate = 100 * people_fully_vaccinated / people_vaccinated) %>%
rename(
`% First Vaccination` = people_vaccinated,
`% Second Vaccination` = people_fully_vaccinated,
`% Second Vaccination out of First` = fully_vaccinated_rate,
Location = location
) %>%
ggplot(aes(`% First Vaccination`, `% Second Vaccination out of First`, size = population, fill = continent, label = Location)) +
geom_hline(yintercept = 100, lty = 2, color = "grey50") +
geom_point(alpha = 0.7, shape = 21) +
theme(legend.position = "none") +
scale_size_continuous(range = c(2,8))
ggplotly(g3,
tooltip = c("Location", '% First Vaccination', '% Second Vaccination out of First'))
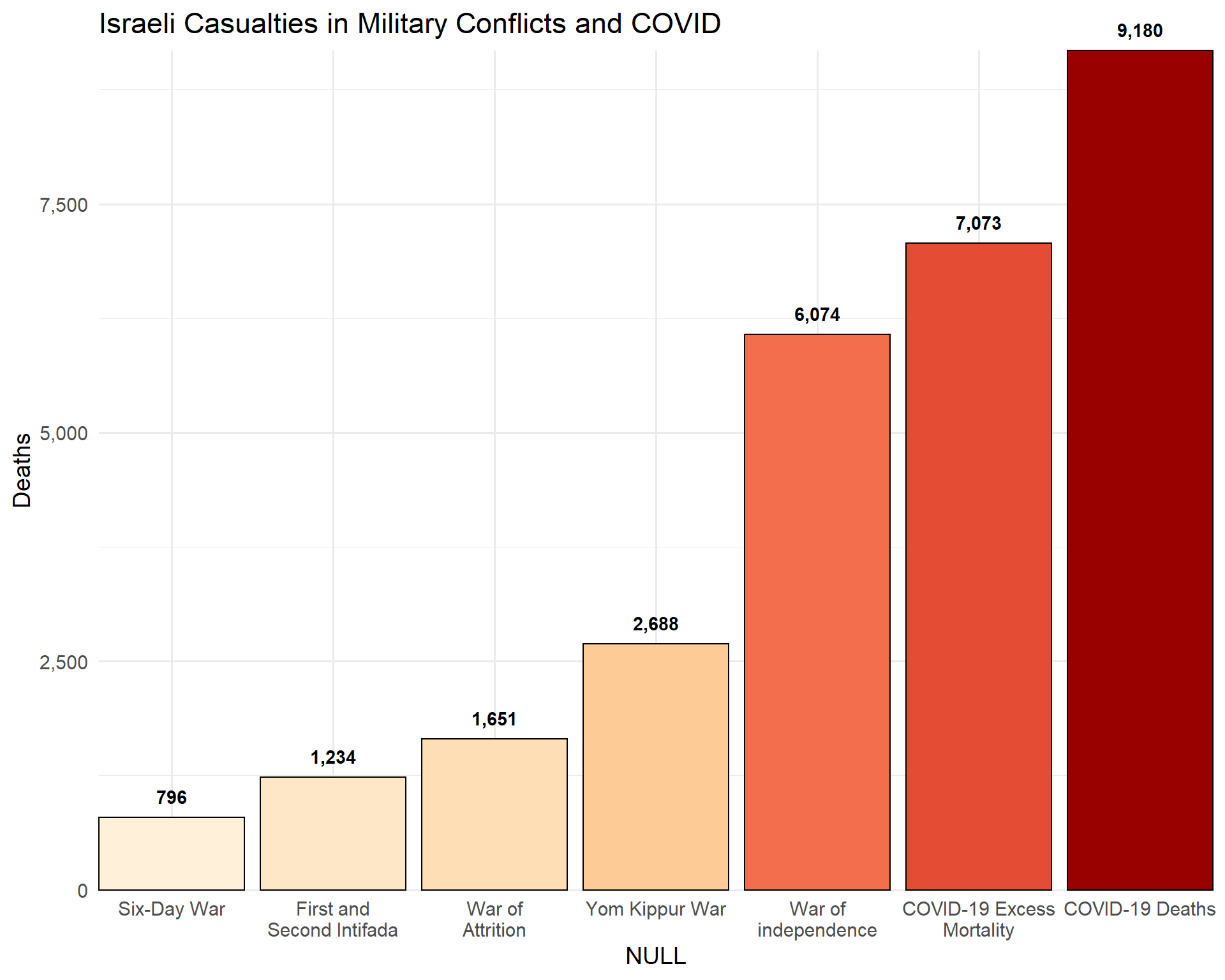
Casualties of military conflicts and of COVID
c19em <- covid %>% filter(location == "Israel") %>%
summarize(max(excess_mortality_cumulative_absolute, na.rm = T)) %>%
pull() %>% round()
c19d <- covid %>% filter(location == "Israel") %>%
summarize(max(total_deaths, na.rm = T)) %>%
pull() %>% round()
conflicts_il <- tribble(
~conflict, ~losses,
"War of independence", 4074 + 2000,
"Six-Day War", 776 + 20,
"War of Attrition", 1424 + 227,
"First and Second Intifada", 301 + 773 + 60 + 100,
"Yom Kippur War", 2688,
"COVID-19 Excess Mortality", c19em,
"COVID-19 Deaths", c19d
)
conflicts_il %>%
mutate(conflict = fct_reorder(str_wrap(conflict, 15), losses)) %>%
ggplot(aes(conflict, losses, fill = losses, label = comma(losses))) +
geom_col(color = "black") +
geom_text(vjust = -1, fontface = "bold") +
scale_fill_distiller(palette = 8, direction = +1) +
coord_cartesian(expand = F, clip = "off") +
labs(title = "Israeli Casualties in Military Conflicts and COVID",
x = "NULL", y = "Deaths") +
theme(legend.position = "none") +
scale_y_continuous(labels = comma_format())
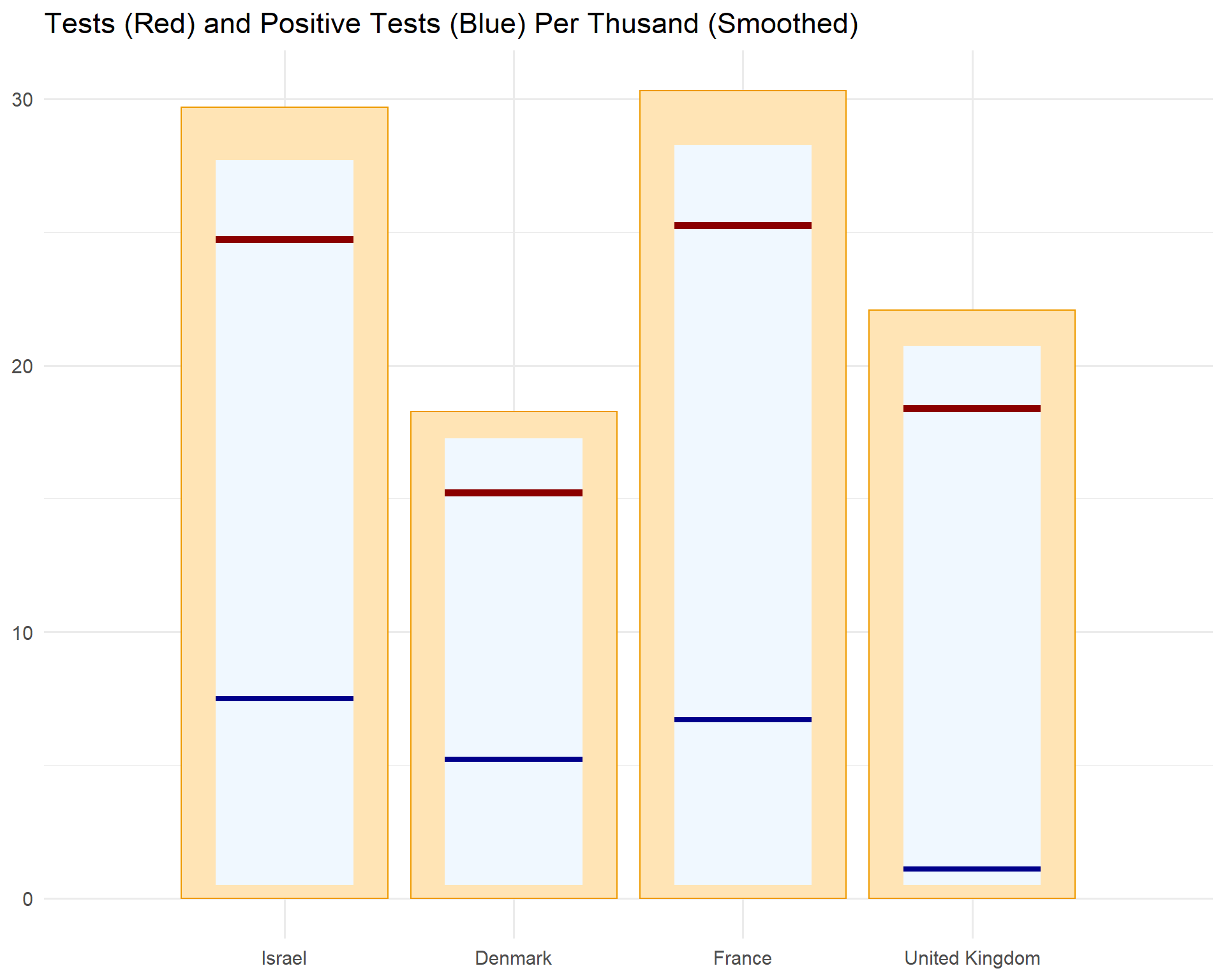
Tests
locs <- c("Israel", "Denmark", "France", "United Kingdom")
covid %>%
select(location,
date,
positive_rate,
new_tests_smoothed_per_thousand) %>%
drop_na(positive_rate, new_tests_smoothed_per_thousand) %>%
filter(location %in% locs) %>%
mutate(location = factor(location, levels = locs)) %>%
group_by(location) %>%
filter(date == max(date, na.rm = T)) %>%
ungroup() %>%
mutate(positive_rate_rate = positive_rate * new_tests_smoothed_per_thousand) %>%
ggplot(aes(x = 1:4, y = new_tests_smoothed_per_thousand)) +
geom_col(aes(y = 1.2 * new_tests_smoothed_per_thousand),
fill = "moccasin",
col = "orange2") +
geom_tile(
aes(
x = 1:4,
width = 0.6,
y = new_tests_smoothed_per_thousand*1.1 / 2 + 0.5,
height = new_tests_smoothed_per_thousand * 1.1
), fill = "aliceblue"
) +
geom_segment(
aes(
x = 1:4 - 0.3,
xend = 1:4 + 0.3,
yend = new_tests_smoothed_per_thousand
),
col = "darkred",
size = 2
) +
geom_segment(
aes(
x = 1:4 - 0.3,
xend = 1:4 + 0.3,
y = positive_rate_rate,
yend = positive_rate_rate
),
col = "darkblue",
size = 1.5
) +
scale_x_discrete(limits = 1:4, labels = locs) +
labs(x = NULL, y = NULL,
title = "Tests (Red) and Positive Tests (Blue) Per Thusand (Smoothed)") +
scale_y_continuous()